In many cropping systems plant-induced modifications to the rhizosphere microbiome promote a community that diminishes productivity and results in reduced yields. Apple Replant Disease (ARD), which was the focus of the joint study, is a model system for the study of soil-borne disease syndromes. The management practice most commonly used against ARD is based on broad-spectrum chemical solutions with drawbacks inherent to their use including potential hazards to the environment and to human health. Soil amendment-based strategies, in particular Brassica seed meal formulations, have a greater long-term, persistent effect than less discriminate management approaches.
However, despite the obvious advantages of seed-meal based sustainable approaches, disease suppression relies on the specific interaction formed between the particular seed meal and the genotype of the apple rootstock. To date, the factors promoting effective versus ineffective treatments are largely unknown. The understanding of microbial community function, in terms of both deleterious and beneficial plant interactions, is the key to finding an effective and persistent solution for management of soil-borne diseases.
The research methodology for this project was based on applying network approaches for the analysis of metagenomics data. Shotgun metagenomic analysis was conducted with data from ‘sick’ vs ‘soil-amended (recovered)’ rhizosphere soil microbiomes. The data was then converted into community-level metabolic networks. Simulations of metabolic interactions examined the functional contribution of treatment-associated taxonomic groups and linked them with specific amendment-induced metabolites. The simulations led to the formulation of predictions for conditions that promoted positive groups and suppressed negative groups. These predictions will enable future research designed to test specific compounds for disease control potential.
“This research demonstrates how genomic-based algorithms can be used to formulate testable hypotheses for strategically engineering the rhizosphere microbiome by identifying specific compounds, which may act as selective modulators of microbial communities. Applying this framework to reduce unpredictable elements in amendment-based solutions promotes the development of ecologically-sound methods for re-establishing a functional microbiome in agro and other ecosystems.”
Berihu, M., Somera, T.S., Malik, A. et al. A framework for the targeted recruitment of crop-beneficial soil taxa based on network analysis of metagenomics data. Microbiome 11, 8 (2023). https://doi.org/10.1186/s40168-022-01438-1
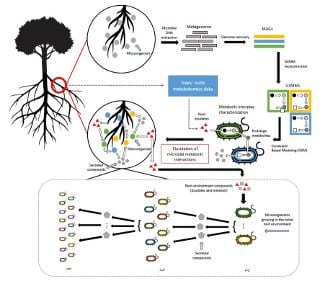
The researchers believe that the outcomes of this project have the potential to contribute to a reduction in the use of chemicals and active use of the indigenous microbiome to safely manage challenges in crop production systems.
This work can serve as a tool for not only understanding the trophic network in soil but also the effects of different environmental factors (such as soil amendments, root exudates) and serve the design of beneficial root communities.








